Unravelling an anticancer drug delivery system in silico
Published on February 14, 2024
Team Kvantify
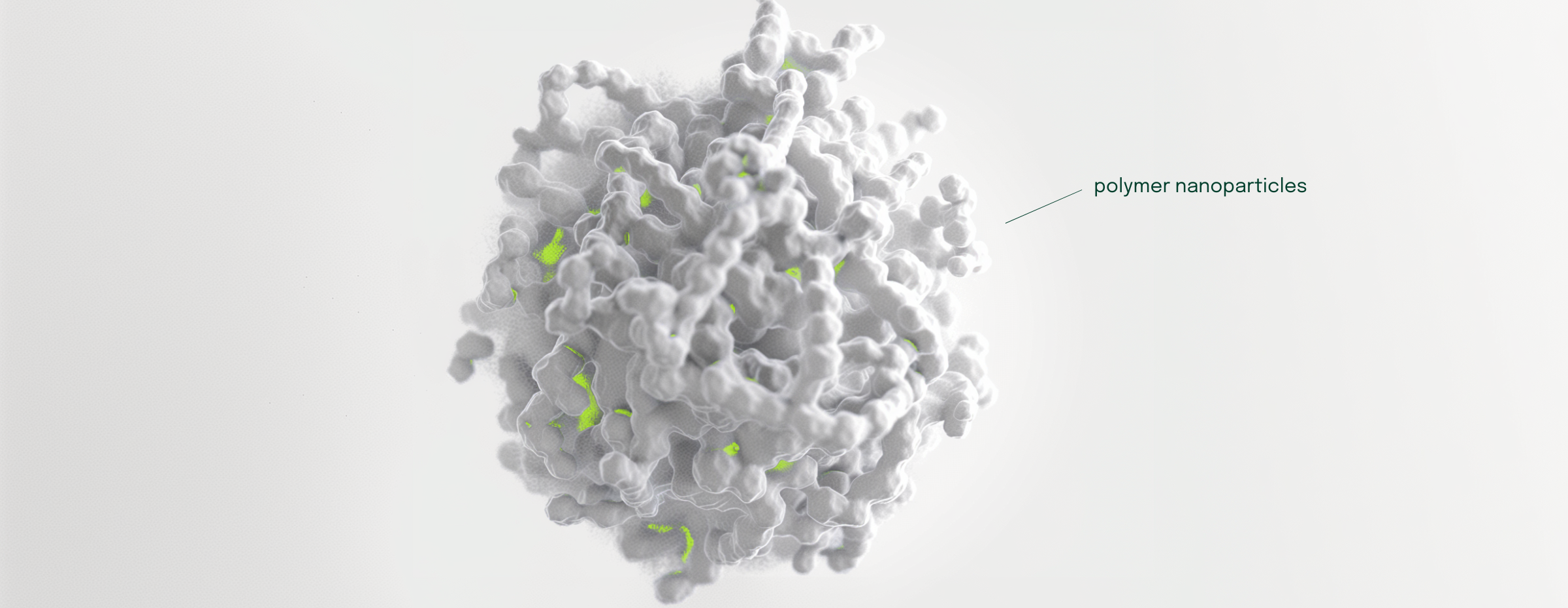
In this blog post, I will present a recent publication in the American Chemical Society’s Nano Letters that I have co-authored with researchers from King’s College London in the related field of drug delivery. Drug delivery is a thriving area of modern pharmaceutical research that is interested in the journey of a drug to its biological target. While we mostly think of the drug molecule in isolation, it can also be incorporated into a formulation containing other chemical matter. For example, drugs can be loaded within nanoparticles made up of lipids, which make up our cell membranes, or polymers, which are long chains of repeating chemical units. An important application of lipid nanoparticles in drug delivery were those used to encapsulate mRNA in COVID-19 vaccines.
Recent experimental results from the group of Prof. Martin Ulmschneider, who is also a coauthor on the paper, have highlighted the anticancer properties of a peptide called EEK when encapsulated in polyethylene glycol-poly (lactic-co-glycolic) acid (PEG–PLGA) nanoparticles. We wanted to better understand what this nanoparticle formulation looks like at the molecular level, so we turned to molecular dynamics simulations as our computational microscope.
We analyzed the molecular simulation and determined that not all EEK peptides are found in similar location within the nanoparticle. Instead, we found that some EEK peptides are found at the interface of the nanoparticle with the surrounding water and some remaining EEK peptides are found deep in the core of the nanoparticle. To understand why this happens, we turned to unsupervised machine learning methods and found the polymers that are in close proximity to the EEK peptides take on different conformations (three-dimensional shapes) to promote the encapsulation of EEK in these different locations within the nanoparticle. Professor Chris Lorenz, who supervised the work, commented that:
“The results presented in this manuscript suggest that having the capability to tune the conformational distribution of the constituent polymers to the chemistry of the delivered therapeutic may be yet another key aspect of the rational design of polymeric drug delivery vehicles.”
This work highlights that molecular scale simulations are uncovering new levels of understanding in ever more complex chemical systems related to drug discovery and drug delivery. It is a pleasure to continue my collaboration with researchers at King’s College London with the support from Kvantify to further explore and broaden my scientific interests.
Discover the full paper here: Therapeutic Peptides Are Preferentially Solubilized in Specific Microenvironments within PEG–PLGA Polymer Nanoparticles | Nano Letters (acs.org)